Hippo: Difference between revisions
→References: SWINK |
|||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
'''Hippo''' is a software package for simulation and analysis of bio-molecules at an atomic level. It has been specifically developed for very efficient protein folding studies in aqueous and membrane environments. The code is very fast due to optimized and hand-coded assembly routines which make use of fast multi-media instructions on modern x86 cpus. Hippo is (partially) parallelized (using industry-standard [http://www.openmp.org/ | [[Image:Gbim.jpg|frame|right|A peptide folds and inserts into a membrane; the membrane is represented by an implicit Generalized Born model.]] | ||
'''Hippo''' is a software package for simulation and analysis of bio-molecules at an atomic level. It has been specifically developed for very efficient protein folding studies in aqueous and membrane environments. The code is very fast due to optimized and hand-coded assembly routines which make use of fast multi-media instructions on modern x86 cpus. Hippo is (partially) parallelized (using industry-standard [http://www.openmp.org/ OpenMP]). | |||
== Features == | == Features == | ||
[[Image:Hippopotamus.jpg|thumb|right|Not this hippo.]] | |||
=== Simulation methods === | === Simulation methods === | ||
* [[Molecular dynamics]] (MD) in NVT, NPT, NVE ensembles | * [[Molecular dynamics]] (MD) in NVT, NPT, NVE ensembles <cite>Proteins2008</cite> | ||
* Metropolis [[Monte Carlo]] (MC) in NVT and NPT ensembles <cite>JACS2004,JPhysChemB2006</cite> | * Metropolis [[Monte Carlo]] (MC) in NVT and NPT ensembles <cite>JACS2004,JPhysChemB2006</cite> | ||
* [[Replica exchange]] with MD and MC | * [[Replica exchange]] with MD and MC <cite>Proteins2007</cite> | ||
=== Force fields === | === Force fields === | ||
| Line 18: | Line 16: | ||
* explicit solvent (water: TIP4P <cite>Jorgensen1985</cite>, SPC <cite>Berendsen1981</cite>) | * explicit solvent (water: TIP4P <cite>Jorgensen1985</cite>, SPC <cite>Berendsen1981</cite>) | ||
* Generalized Born implicit solvent (GB/SA) | * Generalized Born implicit solvent (GB/SA) | ||
* Generalized Born implicit membrane (GB/IM) <cite>Proteins2007,BJ2007,Proteins2005</cite> | * Generalized Born implicit membrane (GB/IM) <cite>Proteins2008,Proteins2007,BJ2007,Proteins2005</cite> | ||
=== Enhanced productivity === | === Enhanced productivity === | ||
| Line 24: | Line 22: | ||
* seamless restarts | * seamless restarts | ||
* intelligent pdb structure loader: reads most pdbs, can complete missing atoms, and builds the topology | * intelligent pdb structure loader: reads most pdbs, can complete missing atoms, and builds the topology | ||
* graphical frontend under development (Windows only) | |||
=== Analysis === | === Analysis === | ||
[[Image:Peptidescan.jpg|frame|right|Adiabatic Generalized Born energy surface for different positions and orientations of a helical peptide in a membrane (output from the ''translation+rotation energy scan'' analysis tool).]] | |||
A growing number of analysis tools are built into Hippo, for instance | A growing number of analysis tools are built into Hippo, for instance | ||
; adiabatic translation+rotation energy scan: determines the Generalized Born energy of a peptide in a membrane <cite>MMB2008,BJ2006</cite>; this allows to decide if a (typically helical) peptide inserts into the membrane and at which depth and angle or if it prefers a surface-bound or even a fully solvated state | |||
* | ; RMSD: calculates the [[RMSD|root mean square deviation]] of trajectory frames to a reference structure | ||
* | ; helicity: degree of helicity of segments along trajectory | ||
; Z - tilt - kink graph: Calculates center of mass, tilt angle, and kink angle of a peptide in a membrane as a function of simulation time. The membrane is in the xy plane, with z = 0 the membrane center. Kink angle is with respect to the membrane normal. <cite>Cordes2002</cite> | |||
; cluster: Performs a cluster analysis using the pairwise method by Daura et al.<cite>Daura1999</cite> | |||
; fit to phase: recenters trajectory on the centre of mass of a phase (such as a lipid membrane) | |||
; fit solute to previous frames: Generate a PDB movie by RMSD fitting the solute of each frame to the previous frame. | |||
The Hippo output format is a binary [[File formats#xyz-movie|xyz-movie]] (see the [http://openbabel.org/wiki/XYZ definition of the xyz format]); thus many other [[analysis]] tools can also be used. | |||
== Installation == | |||
=== Download === | |||
Binaries are publicly available from the [http://www.biowerkzeug.com/ Biowerkzeg.com download page] for '''Linux''' and '''Windows'''. | |||
<!-- , and '''Mac OS X (Intel platform only)'''. --> | |||
=== Installing the software === | |||
Unzip the downloaded file. It will unpack into its own <tt>hippo</tt> directory where you will find | |||
* compiled executables (see below) | |||
** <tt>hippo</tt>: the [[#Simulation methods|MD and MC program]] | |||
** <tt>analyse</tt>: the [[#Analysis|analysis program]] | |||
* the manual ([http://www.biowerkzeug.de/downloads/hippo/hippo_manual.pdf pdf]) | |||
* the [[Hippo license|license]] (to which you consent by downloading) | |||
* the OPLS-AA forcefield file (in Hippo format) | |||
* the readme.txt file | |||
* the <tt>testjobs</tt> directory with example systems | |||
{| class="wikitable" align="right" | |||
|+Hippo executables | |||
!cpu !!Linux !!Windows | |||
|- | |||
|SSSE3 (Core Duo,...) ||<tt>hippo</tt> ||<tt>hippo.exe</tt> | |||
|- | |||
|SSE (PIII,P4,Athlon,...) ||<tt>hippo_p3</tt> ||<tt>hippo_p3.exe</tt> | |||
|} | |||
The package includes binaries that run under Linux and Windows on any Intel or AMD processor that has the SSE or SSSE3 multi media instructions. Depending on your system and needs, choose an executable from the package as shown in the table ''Hippo executables''. | |||
If in doubt, simply try them out in order; if it will not run you will receive an error message such as | |||
Fatal Error: This program was not built to run on the processor in your system. | |||
The allowed processors are: Intel(R) Core(TM) Duo processors and compatible Intel processors | |||
with supplemental Streaming SIMD Extensions 3 (SSSE3) instruction support. | |||
In this case try the <tt>hippo_p3</tt> or <tt>hippo_p3.exe</tt> executable. If this still doesn't work, [http://forums.biowerkzeug.org/viewforum.php?f=6 post a request in the Hippo Installation forum]. | |||
Executables with the <tt>_mpi</tt> extension have been compiled with MPI support ([http://www.mcs.anl.gov/research/projects/mpich2/index.php mpich2]) and are only used for [[replica exchange simulations]] (REXMD). If you just want to run multithreaded simulations just use the standard binaries (which are all compiled with [http://www.openmp.org/ OpenMP]). | |||
=== Test cases === | |||
The <tt>testjobs</tt> directory contains a number of testcases. | |||
Run the <tt>calc_testjobs_linux.bat</tt> or <tt>calc_testjobs_win32.bat</tt> script in order to perform ''all'' tests. On modern processors this should take between 2 and 4 Minutes. | |||
Use the tests in order to get started in running your own systems. | |||
== References == | == References == | ||
| Line 42: | Line 90: | ||
#Proteins2005 pmid=15723347 | #Proteins2005 pmid=15723347 | ||
#JACS2004 pmid=14871118 | #JACS2004 pmid=14871118 | ||
#Kaminski2001 | #Kaminski2001 George A. Kaminski, Richard A. Friesner, Julian Tirado-Rives, and William L. Jorgensen. ''Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides''. J. Phys. Chem. B, 105(28):6474–6487, 2001. [http://dx.doi.org/10.1021/jp003919d 10.1021/jp003919d]. | ||
#Jorgensen1996 W. | #Jorgensen1996 W. L. Jorgensen, D. S. Maxwell, and J. Tirado-Rives. ''Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids''. J. Am. Chem. Soc., 118(45):11225–11236, 1996. [http://dx.doi.org/10.1021/ja9621760 10.1021/ja9621760]. | ||
#Jorgensen1985 W. | #Jorgensen1985 W. L. Jorgensen and J. D. Madura. ''Temperature and size dependence for Monte-Carlo simulations of TIP4P water''. Mol. Phys., 56(6):1381–1392, December 1985. | ||
#Berendsen1981 H. | #Berendsen1981 H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. ''Interaction models for water in relation to protein hydration''. In B. Pullman, editor, Intermolecular Forces, page 331. D. Reidel Publishing Company, Dordrecht, Holland, 1981. | ||
#Cordes2002 pmid=12417206 | #Cordes2002 pmid=12417206 | ||
#Daura1999 X Daura, K Gademann, B Jaun, D Seebach, WF van Gunsteren, and AE Mark. ''Peptide folding: When simulation meets experiment''. Angewandte Chemie-International Edition, 38 (1-2):236–240, 1999. [http://dx.doi.org/10.1002/(SICI)1521-3773(19990115)38:1/2%3C236::AID-ANIE236%3E3.0.CO;2-M <nowiki>10.1002/(SICI)1521-3773(19990115)38:1/2<236::AID-ANIE236>3.0.CO;2-M</nowiki>]. | |||
#Proteins2008 Jakob P. Ulmschneider and Martin B. Ulmschneider. ''Sampling efficiency in explicit and implicit membrane environments studied by peptide folding simulations''. Proteins [http://dx.doi.org/10.1002/prot.22270 10.1002/prot.22270]. | |||
</biblio> | </biblio> | ||
[[Category:Hippo]] | |||
[[Category:Software]] | |||
Latest revision as of 13:49, 13 November 2008
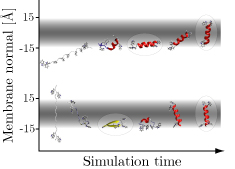
Hippo is a software package for simulation and analysis of bio-molecules at an atomic level. It has been specifically developed for very efficient protein folding studies in aqueous and membrane environments. The code is very fast due to optimized and hand-coded assembly routines which make use of fast multi-media instructions on modern x86 cpus. Hippo is (partially) parallelized (using industry-standard OpenMP).
Features

Simulation methods
- Molecular dynamics (MD) in NVT, NPT, NVE ensembles Proteins2008
- Metropolis Monte Carlo (MC) in NVT and NPT ensembles JACS2004,JPhysChemB2006
- Replica exchange with MD and MC Proteins2007
Force fields
- OPLS-AA Jorgensen1996,Kaminski2001
Solvation models
- explicit solvent (water: TIP4P Jorgensen1985, SPC Berendsen1981)
- Generalized Born implicit solvent (GB/SA)
- Generalized Born implicit membrane (GB/IM) Proteins2008,Proteins2007,BJ2007,Proteins2005
Enhanced productivity
A number of features make it easy to use Hippo so that one can spend more time on working on problems and less time on setting up structures or dealing with system crashes:
- seamless restarts
- intelligent pdb structure loader: reads most pdbs, can complete missing atoms, and builds the topology
- graphical frontend under development (Windows only)
Analysis
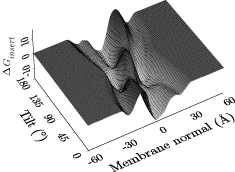
A growing number of analysis tools are built into Hippo, for instance
- adiabatic translation+rotation energy scan
- determines the Generalized Born energy of a peptide in a membrane MMB2008,BJ2006; this allows to decide if a (typically helical) peptide inserts into the membrane and at which depth and angle or if it prefers a surface-bound or even a fully solvated state
- RMSD
- calculates the root mean square deviation of trajectory frames to a reference structure
- helicity
- degree of helicity of segments along trajectory
- Z - tilt - kink graph
- Calculates center of mass, tilt angle, and kink angle of a peptide in a membrane as a function of simulation time. The membrane is in the xy plane, with z = 0 the membrane center. Kink angle is with respect to the membrane normal. Cordes2002
- cluster
- Performs a cluster analysis using the pairwise method by Daura et al.Daura1999
- fit to phase
- recenters trajectory on the centre of mass of a phase (such as a lipid membrane)
- fit solute to previous frames
- Generate a PDB movie by RMSD fitting the solute of each frame to the previous frame.
The Hippo output format is a binary xyz-movie (see the definition of the xyz format); thus many other analysis tools can also be used.
Installation
Download
Binaries are publicly available from the Biowerkzeg.com download page for Linux and Windows.
Installing the software
Unzip the downloaded file. It will unpack into its own hippo directory where you will find
- compiled executables (see below)
- hippo: the MD and MC program
- analyse: the analysis program
- the manual (pdf)
- the license (to which you consent by downloading)
- the OPLS-AA forcefield file (in Hippo format)
- the readme.txt file
- the testjobs directory with example systems
| cpu | Linux | Windows |
|---|---|---|
| SSSE3 (Core Duo,...) | hippo | hippo.exe |
| SSE (PIII,P4,Athlon,...) | hippo_p3 | hippo_p3.exe |
The package includes binaries that run under Linux and Windows on any Intel or AMD processor that has the SSE or SSSE3 multi media instructions. Depending on your system and needs, choose an executable from the package as shown in the table Hippo executables.
If in doubt, simply try them out in order; if it will not run you will receive an error message such as
Fatal Error: This program was not built to run on the processor in your system. The allowed processors are: Intel(R) Core(TM) Duo processors and compatible Intel processors with supplemental Streaming SIMD Extensions 3 (SSSE3) instruction support.
In this case try the hippo_p3 or hippo_p3.exe executable. If this still doesn't work, post a request in the Hippo Installation forum.
Executables with the _mpi extension have been compiled with MPI support (mpich2) and are only used for replica exchange simulations (REXMD). If you just want to run multithreaded simulations just use the standard binaries (which are all compiled with OpenMP).
Test cases
The testjobs directory contains a number of testcases.
Run the calc_testjobs_linux.bat or calc_testjobs_win32.bat script in order to perform all tests. On modern processors this should take between 2 and 4 Minutes.
Use the tests in order to get started in running your own systems.
References
<biblio>
- MMB2008 pmid=18428040
- Proteins2007 pmid=17600830
- BJ2007 pmid=17218457
- BJ2006 pmid=16339877
- JPhysChemB2006 pmid=16913813
- Proteins2005 pmid=15723347
- JACS2004 pmid=14871118
- Kaminski2001 George A. Kaminski, Richard A. Friesner, Julian Tirado-Rives, and William L. Jorgensen. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B, 105(28):6474–6487, 2001. 10.1021/jp003919d.
- Jorgensen1996 W. L. Jorgensen, D. S. Maxwell, and J. Tirado-Rives. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc., 118(45):11225–11236, 1996. 10.1021/ja9621760.
- Jorgensen1985 W. L. Jorgensen and J. D. Madura. Temperature and size dependence for Monte-Carlo simulations of TIP4P water. Mol. Phys., 56(6):1381–1392, December 1985.
- Berendsen1981 H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. Interaction models for water in relation to protein hydration. In B. Pullman, editor, Intermolecular Forces, page 331. D. Reidel Publishing Company, Dordrecht, Holland, 1981.
- Cordes2002 pmid=12417206
- Daura1999 X Daura, K Gademann, B Jaun, D Seebach, WF van Gunsteren, and AE Mark. Peptide folding: When simulation meets experiment. Angewandte Chemie-International Edition, 38 (1-2):236–240, 1999. 10.1002/(SICI)1521-3773(19990115)38:1/2<236::AID-ANIE236>3.0.CO;2-M.
- Proteins2008 Jakob P. Ulmschneider and Martin B. Ulmschneider. Sampling efficiency in explicit and implicit membrane environments studied by peptide folding simulations. Proteins 10.1002/prot.22270.
</biblio>